Regulatory sequence
A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the expression of specific genes within an organism. Regulation of gene expression is an essential feature of all living organisms and viruses.
Description
[edit]
In DNA, regulation of gene expression normally happens at the level of RNA biosynthesis (transcription). It is accomplished through the sequence-specific binding of proteins (transcription factors) that activate or inhibit transcription. Transcription factors may act as activators, repressors, or both. Repressors often act by preventing RNA polymerase from forming a productive complex with the transcriptional initiation region (promoter), while activators facilitate formation of a productive complex. Furthermore, DNA motifs have been shown to be predictive of epigenomic modifications, suggesting that transcription factors play a role in regulating the epigenome.[2]

In RNA, regulation may occur at the level of protein biosynthesis (translation), RNA cleavage, RNA splicing, or transcriptional termination. Regulatory sequences are frequently associated with messenger RNA (mRNA) molecules, where they are used to control mRNA biogenesis or translation. A variety of biological molecules may bind to the RNA to accomplish this regulation, including proteins (e.g., translational repressors and splicing factors), other RNA molecules (e.g., miRNA) and small molecules, in the case of riboswitches.
Activation and implementation
[edit]A regulatory DNA sequence does not regulate unless it is activated. Different regulatory sequences are activated and then implement their regulation by different mechanisms.
Enhancer activation and implementation
[edit]Expression of genes in mammals can be upregulated when signals are transmitted to the promoters associated with the genes. Cis-regulatory DNA sequences that are located in DNA regions distant from the promoters of genes can have very large effects on gene expression, with some genes undergoing up to 100-fold increased expression due to such a cis-regulatory sequence.[3] These cis-regulatory sequences include enhancers, silencers, insulators and tethering elements.[4] Among this constellation of sequences, enhancers and their associated transcription factor proteins have a leading role in the regulation of gene expression.[5]
Enhancers are sequences of the genome that are major gene-regulatory elements. Enhancers control cell-type-specific gene expression programs, most often by looping through long distances to come in physical proximity with the promoters of their target genes.[6] In a study of brain cortical neurons, 24,937 loops were found, bringing enhancers to promoters.[3] Multiple enhancers, each often at tens or hundreds of thousands of nucleotides distant from their target genes, loop to their target gene promoters and coordinate with each other to control expression of their common target gene.[6]
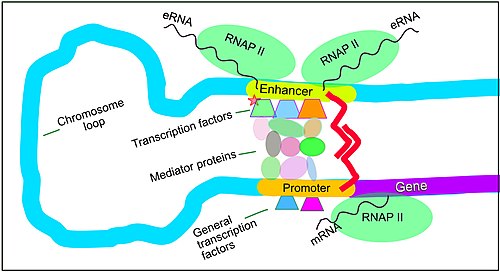
The schematic illustration in this section shows an enhancer looping around to come into close physical proximity with the promoter of a target gene. The loop is stabilized by a dimer of a connector protein (e.g. dimer of CTCF or YY1), with one member of the dimer anchored to its binding motif on the enhancer and the other member anchored to its binding motif on the promoter (represented by the red zigzags in the illustration).[7] Several cell function specific transcription factor proteins (in 2018 Lambert et al. indicated there were about 1,600 transcription factors in a human cell[8]) generally bind to specific motifs on an enhancer[9] and a small combination of these enhancer-bound transcription factors, when brought close to a promoter by a DNA loop, govern the level of transcription of the target gene. Mediator (coactivator) (a complex usually consisting of about 26 proteins in an interacting structure) communicates regulatory signals from enhancer DNA-bound transcription factors directly to the RNA polymerase II (RNAP II) enzyme bound to the promoter.[10]
Enhancers, when active, are generally transcribed from both strands of DNA with RNA polymerases acting in two different directions, producing two eRNAs as illustrated in the Figure.[11] An inactive enhancer may be bound by an inactive transcription factor. Phosphorylation of the transcription factor may activate it and that activated transcription factor may then activate the enhancer to which it is bound (see small red star representing phosphorylation of a transcription factor bound to an enhancer in the illustration).[12] An activated enhancer begins transcription of its RNA before activating a promoter to initiate transcription of messenger RNA from its target gene.[13]
Transcription factor binding sites within enhancers (see figure above) are usually about 10 base pairs long, though they can vary from just a few to about 20 base pairs.[14] Enhancers usually have about 10 transcription factor binding sites within an average enhancer site of about 204 base pairs.[15] Examining enhancer-gene regulatory interactions occurring in 352 cell types and tissues, more than 13 million active enhancers were found.[16]
Super-enhancer
[edit]
While enhancers are needed for transcription of genes in a cell above low levels, a cluster of enhancers, known as a super-enhancer, can cause transcription of a target gene at even higher levels. Super-enhancers usually drive genes needed for cell identity to express at high levels.[17][18] In cancers, a super-enhancer may also drive a particular oncogene to express at a high level.[17][18]
A super-enhancer is defined as a cluster of typical enhancers in close genomic proximity (within about 9,000[17] to 22,0000[19] base pairs in length) that, all together, regulate the expression of a target gene.[20] Super-enhancer-driven genes are expressed at significantly higher levels than the expression of genes under the control of typical enhancers.[20]
A diagram of a super-enhancer is shown in the Figure in this section. In this Figure, the super-enhancer is 12,000 nucleotides long and has four typical enhancers within its length. Each of the typical enhancers simultaneously contacts the promoter region of the same target gene. Each typical enhancer within the super-enhancer has multiple DNA motifs to which transcription factors bind. Each typical enhancer is also bound to a 26-component mediator complex which transmits the signals from the transcription factors bound to the enhancer to the promoter of their joint target gene. The protein BRD4 forms a complex with each typical enhancer in the super-enhancer and helps to stabilizes the super-enhancer structure.[21] In addition, the architectural protein YY1 (indicated by paired red zigzags) helps keep the loops together that bring the typical enhancers to their target gene in the super-enhancer.[7] Therefore, there are many proteins in close association at a super-enhancer. These proteins generally have a structured domain as well as a tail with an intrinsically disordered region (IDR).[22] Many of the IDRs of these proteins interact with each other, thereby forming a water-excluding gel or phase-separated condensate around the super-enhancer.[22]
Some super-enhancers induce very high levels of transcription such as the mouse α-globin super-enhancer[23] and the Wap super-enhancer.[24] The mouse α-globin super-enhancer has five typical enhancers within the super-enhancer. Only when acting together, they increase transcription of the α-globin gene by 450-fold.[23] In another example, the mouse Wap super-enhancer includes three typical enhancers. Only when the three typical enhancers act together do they increase transcription of the Wap gene by 1000-fold.[24]
The enhancers within the super-enhancers described above act synergistically. However, in a second type of super-enhancer, the component enhancers act additively. In a third group, super-enhancers appear to act “logistically” where promoter activity reaches a limit. One study examined 773 target genes that were paired with near-by groups of possible super-enhancers (with 2–20 enhancers in close proximity likely acting as super-enhancers). In this study it appeared that 277, 92, and 250 of the likely super-enhancers acted by the additive, synergistic, and logistic models.[25]
Super-enhancers may occupy regions of the genome about 10,000 to 60,000 nucleotides long.[26] while typical enhancers are each about 204 base pairs long.[15] When 8 types of cells were evaluated, super-enhancers constituted between 2.5% to 10.9% of the enhancers driving transcription while typical enhancers were the majority of enhancers driving transcription. There were between 257 and 1,099 super-enhancers in these eight cell types and between 5,512 and 23,869 typical enhancers.[27]
While super-enhancers are only active at about 2.5% – 10.9% of actively transcribed sites in a cell, they recruit transcription machinery more actively than at typical single enhancers. The super-enhancers in a cell utilize about 12% to 36% of the RNA polymerases, mediator proteins, BRD4 proteins, and other transcription machinery of the cell.[17]
CpG island methylation and demethylation
[edit]
5-Methylcytosine (5-mC) is a methylated form of the DNA base cytosine (see figure). 5-mC is an epigenetic marker found predominantly on cytosines within CpG dinucleotides, which consist of a cytosine is followed by a guanine reading in the 5' to 3' direction along the DNA strand (CpG sites). About 28 million CpG dinucleotides occur in the human genome.[28] In most tissues of mammals, on average, 70% to 80% of CpG cytosines are methylated (forming 5-methyl-CpG, or 5-mCpG).[29] Methylated cytosines within CpG sequences often occur in groups, called CpG islands. About 59% of promoter sequences have a CpG island while only about 6% of enhancer sequences have a CpG island.[30] CpG islands constitute regulatory sequences, since if CpG islands are methylated in the promoter of a gene this can reduce or silence gene expression.[31]
DNA methylation regulates gene expression through interaction with methyl binding domain (MBD) proteins, such as MeCP2, MBD1 and MBD2. These MBD proteins bind most strongly to highly methylated CpG islands.[32] These MBD proteins have both a methyl-CpG-binding domain and a transcriptional repression domain.[32] They bind to methylated DNA and guide or direct protein complexes with chromatin remodeling and/or histone modifying activity to methylated CpG islands. MBD proteins generally repress local chromatin by means such as catalyzing the introduction of repressive histone marks or creating an overall repressive chromatin environment through nucleosome remodeling and chromatin reorganization.[32]
Transcription factors are proteins that bind to specific DNA sequences in order to regulate the expression of a given gene. The binding sequence for a transcription factor in DNA is usually about 10 or 11 nucleotides long. There are approximately 1,400 different transcription factors encoded in the human genome, and they constitute about 6% of all human protein coding genes.[33] About 94% of transcription factor binding sites that are associated with signal-responsive genes occur in enhancers while only about 6% of such sites occur in promoters.[9]
EGR1 is a transcription factor important for regulation of methylation of CpG islands. An EGR1 transcription factor binding site is frequently located in enhancer or promoter sequences.[34] There are about 12,000 binding sites for EGR1 in the mammalian genome and about half of EGR1 binding sites are located in promoters and half in enhancers.[34] The binding of EGR1 to its target DNA binding site is insensitive to cytosine methylation in the DNA.[34]
While only small amounts of EGR1 protein are detectable in cells that are un-stimulated, EGR1 translation into protein at one hour after stimulation is markedly elevated.[35] Expression of EGR1 in various types of cells can be stimulated by growth factors, neurotransmitters, hormones, stress and injury.[35] In the brain, when neurons are activated, EGR1 proteins are upregulated, and they bind to (recruit) pre-existing TET1 enzymes, which are highly expressed in neurons. TET enzymes can catalyze demethylation of 5-methylcytosine. When EGR1 transcription factors bring TET1 enzymes to EGR1 binding sites in promoters, the TET enzymes can demethylate the methylated CpG islands at those promoters. Upon demethylation, these promoters can then initiate transcription of their target genes. Hundreds of genes in neurons are differentially expressed after neuron activation through EGR1 recruitment of TET1 to methylated regulatory sequences in their promoters.[34]
Activation by double- or single-strand breaks
[edit]About 600 regulatory sequences in promoters and about 800 regulatory sequences in enhancers appear to depend on double-strand breaks initiated by topoisomerase 2β (TOP2B) for activation.[36][37] The induction of particular double-strand breaks is specific with respect to the inducing signal. When neurons are activated in vitro, just 22 TOP2B-induced double-strand breaks occur in their genomes.[38] However, when contextual fear conditioning is carried out in a mouse, this conditioning causes hundreds of gene-associated DSBs in the medial prefrontal cortex and hippocampus, which are important for learning and memory.[39]

Such TOP2B-induced double-strand breaks are accompanied by at least four enzymes of the non-homologous end joining (NHEJ) DNA repair pathway (DNA-PKcs, KU70, KU80 and DNA LIGASE IV) (see figure). These enzymes repair the double-strand breaks within about 15 minutes to 2 hours.[38][40] The double-strand breaks in the promoter are thus associated with TOP2B and at least these four repair enzymes. These proteins are present simultaneously on a single promoter nucleosome (there are about 147 nucleotides in the DNA sequence wrapped around a single nucleosome) located near the transcription start site of their target gene.[40]
The double-strand break introduced by TOP2B apparently frees the part of the promoter at an RNA polymerase–bound transcription start site to physically move to its associated enhancer. This allows the enhancer, with its bound transcription factors and mediator proteins, to directly interact with the RNA polymerase that had been paused at the transcription start site to start transcription.[38][10]
Similarly, topoisomerase I (TOP1) enzymes appear to be located at many enhancers, and those enhancers become activated when TOP1 introduces a single-strand break.[41] TOP1 causes single-strand breaks in particular enhancer DNA regulatory sequences when signaled by a specific enhancer-binding transcription factor.[41] Topoisomerase I breaks are associated with different DNA repair factors than those surrounding TOP2B breaks. In the case of TOP1, the breaks are associated most immediately with DNA repair enzymes MRE11, RAD50 and ATR.[41]
Examples
[edit]Genomes can be analyzed systematically to identify regulatory regions.[42] Conserved non-coding sequences often contain regulatory regions, and so they are often the subject of these analyses.
- CAAT box
- CCAAT box
- Operator (biology)
- Pribnow box
- TATA box
- SECIS element, mRNA
- Polyadenylation signal, mRNA
- A-box
- Z-box
- C-box
- E-box
- G-box
Insulin gene
[edit]Regulatory sequences for the insulin gene are:[43]
- A5
- Z
- negative regulatory element (NRE)[44]
- C2
- E2
- A3
- cAMP response element
- A2
- CAAT enhancer binding (CEB)
- C1
- E1
- G1
See also
[edit]- Regulator gene
- Regulation of gene expression
- Cis-acting element
- Gene regulatory network
- Open Regulatory Annotation Database
- Operon
- DNA binding site
- Promoter
- Trans-acting factor
- ORegAnno
References
[edit]- ^ Shafee, Thomas; Lowe, Rohan (2017). "Eukaryotic and prokaryotic gene structure". WikiJournal of Medicine. 4 (1). doi:10.15347/wjm/2017.002. ISSN 2002-4436.
- ^ Whitaker JW, Zhao Chen, Wei Wang. (2014) Predicting the Human Epigenome from DNA Motifs. Nature Methods. doi:10.1038/nmeth.3065
- ^ a b Beagan JA, Pastuzyn ED, Fernandez LR, Guo MH, Feng K, Titus KR, et al. (June 2020). "Three-dimensional genome restructuring across timescales of activity-induced neuronal gene expression". Nature Neuroscience. 23 (6): 707–717. doi:10.1038/s41593-020-0634-6. PMC 7558717. PMID 32451484.
- ^ Verheul TC, van Hijfte L, Perenthaler E, Barakat TS (2020). "The Why of YY1: Mechanisms of Transcriptional Regulation by Yin Yang 1". Frontiers in Cell and Developmental Biology. 8: 592164. doi:10.3389/fcell.2020.592164. PMC 7554316. PMID 33102493.
- ^ Spitz F, Furlong EE (September 2012). "Transcription factors: from enhancer binding to developmental control". Nature Reviews. Genetics. 13 (9): 613–26. doi:10.1038/nrg3207. PMID 22868264. S2CID 205485256.
- ^ a b Schoenfelder S, Fraser P (August 2019). "Long-range enhancer-promoter contacts in gene expression control". Nature Reviews. Genetics. 20 (8): 437–455. doi:10.1038/s41576-019-0128-0. PMID 31086298. S2CID 152283312.
- ^ a b Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL, Guo YE, Hnisz D, Jaenisch R, Bradner JE, Gray NS, Young RA (December 2017). "YY1 Is a Structural Regulator of Enhancer-Promoter Loops". Cell. 171 (7): 1573–1588.e28. doi:10.1016/j.cell.2017.11.008. PMC 5785279. PMID 29224777.
- ^ Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. (February 2018). "The Human Transcription Factors". Cell. 172 (4): 650–665. doi:10.1016/j.cell.2018.01.029. PMID 29425488.
- ^ a b Grossman SR, Engreitz J, Ray JP, Nguyen TH, Hacohen N, Lander ES (July 2018). "Positional specificity of different transcription factor classes within enhancers". Proceedings of the National Academy of Sciences of the United States of America. 115 (30): E7222 – E7230. Bibcode:2018PNAS..115E7222G. doi:10.1073/pnas.1804663115. PMC 6065035. PMID 29987030.
- ^ a b Allen BL, Taatjes DJ (March 2015). "The Mediator complex: a central integrator of transcription". Nature Reviews. Molecular Cell Biology. 16 (3): 155–66. doi:10.1038/nrm3951. PMC 4963239. PMID 25693131.
- ^ Mikhaylichenko O, Bondarenko V, Harnett D, Schor IE, Males M, Viales RR, Furlong EE (January 2018). "The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription". Genes & Development. 32 (1): 42–57. doi:10.1101/gad.308619.117. PMC 5828394. PMID 29378788.
- ^ Li QJ, Yang SH, Maeda Y, Sladek FM, Sharrocks AD, Martins-Green M (January 2003). "MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300". The EMBO Journal. 22 (2): 281–91. doi:10.1093/emboj/cdg028. PMC 140103. PMID 12514134.
- ^ Carullo NV, Phillips Iii RA, Simon RC, Soto SA, Hinds JE, Salisbury AJ, et al. (September 2020). "Enhancer RNAs predict enhancer-gene regulatory links and are critical for enhancer function in neuronal systems". Nucleic Acids Research. 48 (17): 9550–9570. doi:10.1093/nar/gkaa671. PMC 7515708. PMID 32810208.
- ^ Wang S, Zhang Q, Shen Z, He Y, Chen ZH, Li J, Huang DS (June 2021). "Predicting transcription factor binding sites using DNA shape features based on shared hybrid deep learning architecture". Mol Ther Nucleic Acids. 24: 154–163. doi:10.1016/j.omtn.2021.02.014. PMC 7972936. PMID 33767912.
- ^ a b Meuleman W, Muratov A, Rynes E, Halow J, Lee K, Bates D, Diegel M, Dunn D, Neri F, Teodosiadis A, Reynolds A, Haugen E, Nelson J, Johnson A, Frerker M, Buckley M, Sandstrom R, Vierstra J, Kaul R, Stamatoyannopoulos J (August 2020). "Index and biological spectrum of human DNase I hypersensitive sites". Nature. 584 (7820): 244–251. Bibcode:2020Natur.584..244M. doi:10.1038/s41586-020-2559-3. PMC 7422677. PMID 32728217.
- ^ Gschwind AR, Mualim KS, Karbalayghareh A, Sheth MU, Dey KK, Jagoda E, Nurtdinov RN, Xi W, Tan AS, Jones H, Ma XR, Yao D, Nasser J, Avsec Ž, James BT, Shamim MS, Durand NC, Rao SS, Mahajan R, Doughty BR, Andreeva K, Ulirsch JC, Fan K, Perez EM, Nguyen TC, Kelley DR, Finucane HK, Moore JE, Weng Z, Kellis M, Bassik MC, Price AL, Beer MA, Guigó R, Stamatoyannopoulos JA, Lieberman Aiden E, Greenleaf WJ, Leslie CS, Steinmetz LM, Kundaje A, Engreitz JM (November 2023). "An encyclopedia of enhancer-gene regulatory interactions in the human genome". bioRxiv. doi:10.1101/2023.11.09.563812. PMC 10680627. PMID 38014075.
- ^ a b c d Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA (November 2013). "Super-enhancers in the control of cell identity and disease". Cell. 155 (4): 934–47. doi:10.1016/j.cell.2013.09.053. PMC 3841062. PMID 24119843.
- ^ a b Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, Bradner JE, Young RA (April 2015). "Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers". Mol Cell. 58 (2): 362–70. doi:10.1016/j.molcel.2015.02.014. PMC 4402134. PMID 25801169.
- ^ Khan A, Mathelier A, Zhang X (2018). "Super-enhancers are transcriptionally more active and cell type-specific than stretch enhancers". Epigenetics. 13 (9): 910–922. doi:10.1080/15592294.2018.1514231. PMC 6284781. PMID 30169995.
- ^ a b Sengupta S, George RE (April 2017). "Super-Enhancer-Driven Transcriptional Dependencies in Cancer". Trends Cancer. 3 (4): 269–281. doi:10.1016/j.trecan.2017.03.006. PMC 5546010. PMID 28718439.
- ^ Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, Li CH, Guo YE, Day DS, Schuijers J, Vasile E, Malik S, Hnisz D, Lee TI, Cisse II, Roeder RG, Sharp PA, Chakraborty AK, Young RA (July 2018). "Coactivator condensation at super-enhancers links phase separation and gene control". Science. 361 (6400). doi:10.1126/science.aar3958. PMC 6092193. PMID 29930091.
- ^ a b Boija A, Klein IA, Sabari BR, Dall'Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, Abraham BJ, Afeyan LK, Guo YE, Rimel JK, Fant CB, Schuijers J, Lee TI, Taatjes DJ, Young RA (December 2018). "Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains". Cell. 175 (7): 1842–1855.e16. doi:10.1016/j.cell.2018.10.042. PMC 6295254. PMID 30449618.
- ^ a b Blayney JW, Francis H, Rampasekova A, Camellato B, Mitchell L, Stolper R, Cornell L, Babbs C, Boeke JD, Higgs DR, Kassouf M (December 2023). "Super-enhancers include classical enhancers and facilitators to fully activate gene expression". Cell. 186 (26): 5826–5839.e18. doi:10.1016/j.cell.2023.11.030. PMC 10858684. PMID 38101409.
- ^ a b Shin HY, Willi M, HyunYoo K, Zeng X, Wang C, Metser G, Hennighausen L (August 2016). "Hierarchy within the mammary STAT5-driven Wap super-enhancer". Nat Genet. 48 (8): 904–911. doi:10.1038/ng.3606. PMC 4963296. PMID 27376239.
- ^ Choi J, Lysakovskaia K, Stik G, Demel C, Söding J, Tian TV, Graf T, Cramer P (March 2021). "Evidence for additive and synergistic action of mammalian enhancers during cell fate determination". eLife. 10. doi:10.7554/eLife.65381. PMC 8004103. PMID 33770473.
- ^ Wang X, Cairns MJ, Yan J (December 2019). "Super-enhancers in transcriptional regulation and genome organization". Nucleic Acids Res. 47 (22): 11481–11496. doi:10.1093/nar/gkz1038. PMC 7145697. PMID 31724731.
- ^ Niederriter AR, Varshney A, Parker SC, Martin DM (November 2015). "Super Enhancers in Cancers, Complex Disease, and Developmental Disorders". Genes (Basel). 6 (4): 1183–200. doi:10.3390/genes6041183. PMC 4690034. PMID 26569311.
- ^ Lövkvist C, Dodd IB, Sneppen K, Haerter JO (June 2016). "DNA methylation in human epigenomes depends on local topology of CpG sites". Nucleic Acids Research. 44 (11): 5123–32. doi:10.1093/nar/gkw124. PMC 4914085. PMID 26932361.
- ^ Jabbari K, Bernardi G (May 2004). "Cytosine methylation and CpG, TpG (CpA) and TpA frequencies". Gene. 333: 143–9. doi:10.1016/j.gene.2004.02.043. PMID 15177689.
- ^ Steinhaus R, Gonzalez T, Seelow D, Robinson PN (June 2020). "Pervasive and CpG-dependent promoter-like characteristics of transcribed enhancers". Nucleic Acids Research. 48 (10): 5306–5317. doi:10.1093/nar/gkaa223. PMC 7261191. PMID 32338759.
- ^ Bird A (January 2002). "DNA methylation patterns and epigenetic memory". Genes & Development. 16 (1): 6–21. doi:10.1101/gad.947102. PMID 11782440.
- ^ a b c Du Q, Luu PL, Stirzaker C, Clark SJ (2015). "Methyl-CpG-binding domain proteins: readers of the epigenome". Epigenomics. 7 (6): 1051–73. doi:10.2217/epi.15.39. PMID 25927341.
- ^ Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM (April 2009). "A census of human transcription factors: function, expression and evolution". Nature Reviews. Genetics. 10 (4): 252–63. doi:10.1038/nrg2538. PMID 19274049. S2CID 3207586.
- ^ a b c d Sun Z, Xu X, He J, Murray A, Sun MA, Wei X, et al. (August 2019). "EGR1 recruits TET1 to shape the brain methylome during development and upon neuronal activity". Nature Communications. 10 (1): 3892. Bibcode:2019NatCo..10.3892S. doi:10.1038/s41467-019-11905-3. PMC 6715719. PMID 31467272.
- ^ a b Kubosaki A, Tomaru Y, Tagami M, Arner E, Miura H, Suzuki T, et al. (2009). "Genome-wide investigation of in vivo EGR-1 binding sites in monocytic differentiation". Genome Biology. 10 (4): R41. doi:10.1186/gb-2009-10-4-r41. PMC 2688932. PMID 19374776.
- ^ Dellino GI, Palluzzi F, Chiariello AM, Piccioni R, Bianco S, Furia L, et al. (June 2019). "Release of paused RNA polymerase II at specific loci favors DNA double-strand-break formation and promotes cancer translocations". Nature Genetics. 51 (6): 1011–1023. doi:10.1038/s41588-019-0421-z. PMID 31110352. S2CID 159041612. Archived from the original on May 25, 2022.
- ^ Singh S, Szlachta K, Manukyan A, Raimer HM, Dinda M, Bekiranov S, Wang YH (March 2020). "Pausing sites of RNA polymerase II on actively transcribed genes are enriched in DNA double-stranded breaks". J Biol Chem. 295 (12): 3990–4000. doi:10.1074/jbc.RA119.011665. PMC 7086017. PMID 32029477.
- ^ a b c Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, et al. (June 2015). "Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes". Cell. 161 (7): 1592–605. doi:10.1016/j.cell.2015.05.032. PMC 4886855. PMID 26052046.
- ^ Stott RT, Kritsky O, Tsai LH (2021). "Profiling DNA break sites and transcriptional changes in response to contextual fear learning". PLOS ONE. 16 (7): e0249691. Bibcode:2021PLoSO..1649691S. doi:10.1371/journal.pone.0249691. PMC 8248687. PMID 34197463.
- ^ a b Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG (June 2006). "A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription". Science. 312 (5781): 1798–802. Bibcode:2006Sci...312.1798J. doi:10.1126/science.1127196. PMID 16794079. S2CID 206508330.
- ^ a b c Puc J, Kozbial P, Li W, Tan Y, Liu Z, Suter T, et al. (January 2015). "Ligand-dependent enhancer activation regulated by topoisomerase-I activity". Cell. 160 (3): 367–80. doi:10.1016/j.cell.2014.12.023. PMC 4422651. PMID 25619691.
- ^ Stepanova M, Tiazhelova T, Skoblov M, Baranova A (May 2005). "A comparative analysis of relative occurrence of transcription factor binding sites in vertebrate genomes and gene promoter areas". Bioinformatics. 21 (9): 1789–96. doi:10.1093/bioinformatics/bti307. PMID 15699025.
- ^ Melloul D, Marshak S, Cerasi E (March 2002). "Regulation of insulin gene transcription". Diabetologia. 45 (3): 309–26. doi:10.1007/s00125-001-0728-y. PMID 11914736.
- ^ Jang WG, Kim EJ, Park KG, Park YB, Choi HS, Kim HJ, et al. (January 2007). "Glucocorticoid receptor mediated repression of human insulin gene expression is regulated by PGC-1alpha". Biochemical and Biophysical Research Communications. 352 (3): 716–21. doi:10.1016/j.bbrc.2006.11.074. PMID 17150186.
